Wheeler Microfluidics Laboratory, Department of Chemistry, Institute of Biomaterials and Biomedical Engineering (IBBME), University of Toronto
Supervisor: Dr. Aaron Wheeler
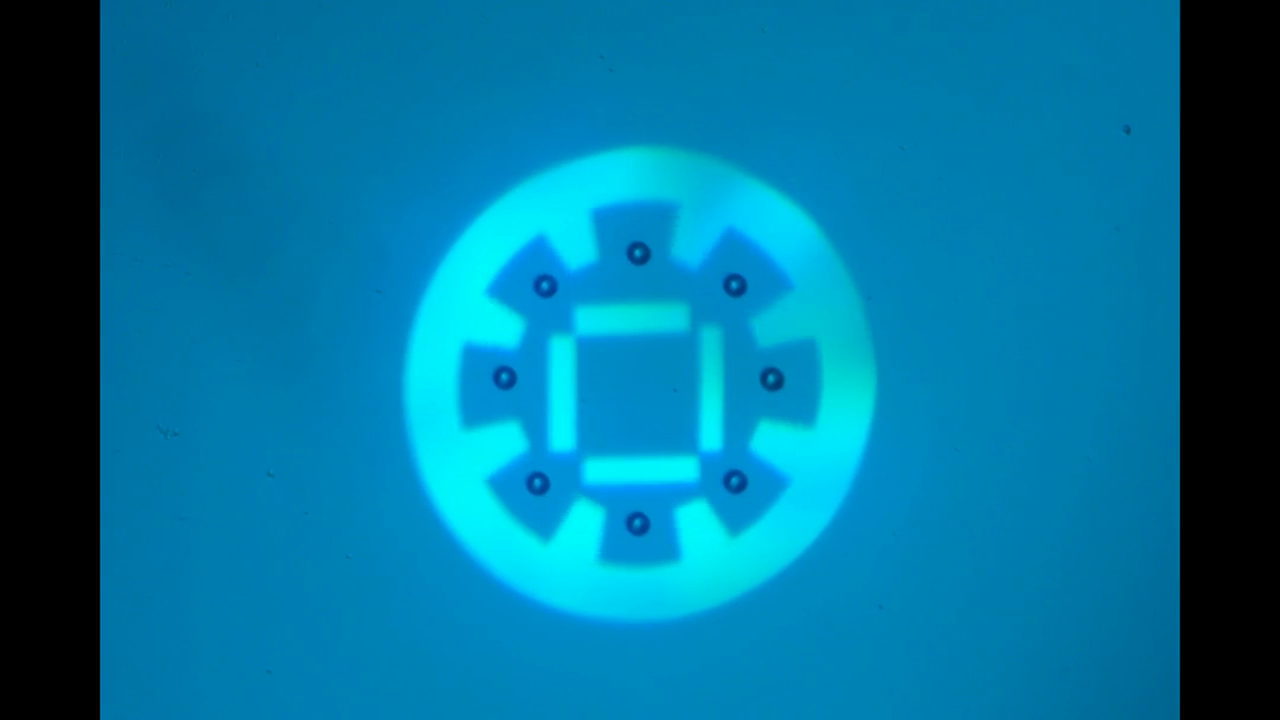
"Manipulate micro-beads with light: roulette, minion and American captain"
This video shows the use of the light pattern to manipulate micro-beads in an optoelectronic tweezer (OET) system. OET is an optical micromanipulation technology that uses light-induced dielectrophoresis force to control and actuate micro-scale objects such as dielectric beads and living cells. In this video, we demonstrate the use of versatile interesting light patterns to move and rotate micro-beads in a highly-controlled manner.
Department of Chemistry, McGill University
Supervisor: Dr. Hanadi Sleiman
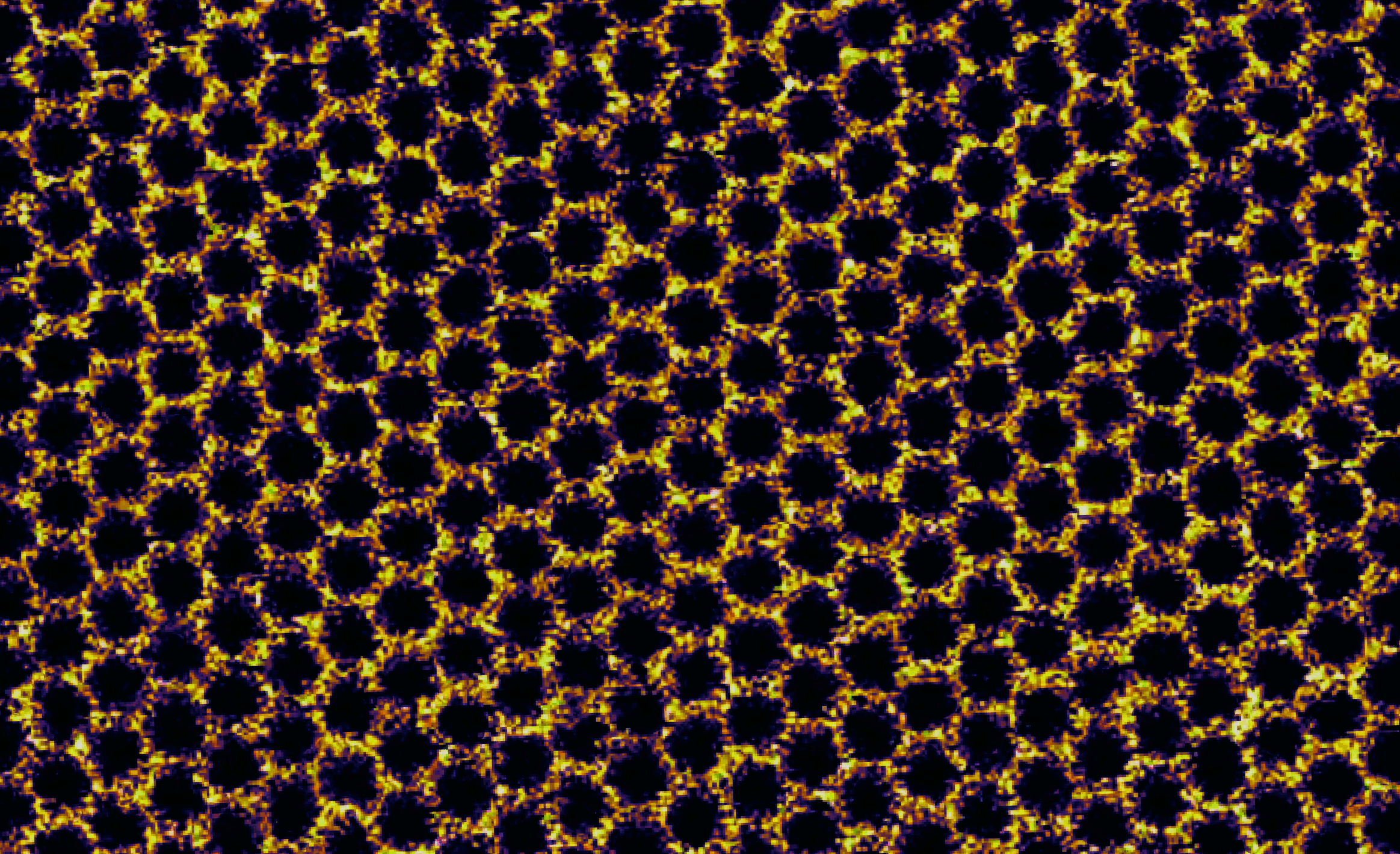
Avakyan, N., Conway, J. W. & Sleiman, H. F. J. Am. Chem. Soc. 139, 12027-12034 (2017)
Copyright 2017 American Chemical Society
"A nanoscale DNA honeycomb"
This honeycomb was not built by bees – it arises from the assembly of Y-shaped tiles made from synthetic DNA strands. Each tile measures only ~10 nanometers across and represents a vertex in the hexagonal pattern. Each also carries a cholesterol molecule that sinks like an anchor into a fluid, cell membrane-like support material composed of lipid molecules (DOPC). On this lipid bilayer support, tiles diffuse and find their neighbours, forming an ordered network by using weak interactions to link them together. Contrary to most long-range assembly in DNA nanotechnology, the contacts between the tiles in this image are mediated by adhesion of blunt-ended duplexes rather than binding of short, complementary single-stranded overhangs (sticky-ends). These weak blunt-end contacts form and break apart easily allowing for network reconfiguration that corrects assembly errors, thus giving rise to long-range order with few defects. Such precise long-range assembly on a biological interface allows us to envisage new developments in nanotechnology, from the organization of protein complexes into specific patterns for sensing applications to artificial light-harvesting platforms. This image of sample topography was obtained by high-resolution atomic force microscopy (AFM) on a Bruker MultiMode 8 instrument equipped with a Nanoscope 5 controller. Imaging was carried out under ambient conditions in fluid using the PeakForce Tapping mode.
Department of Materials Science & Engineering, McMaster University
Supervisor: Dr. Nabil Bassim
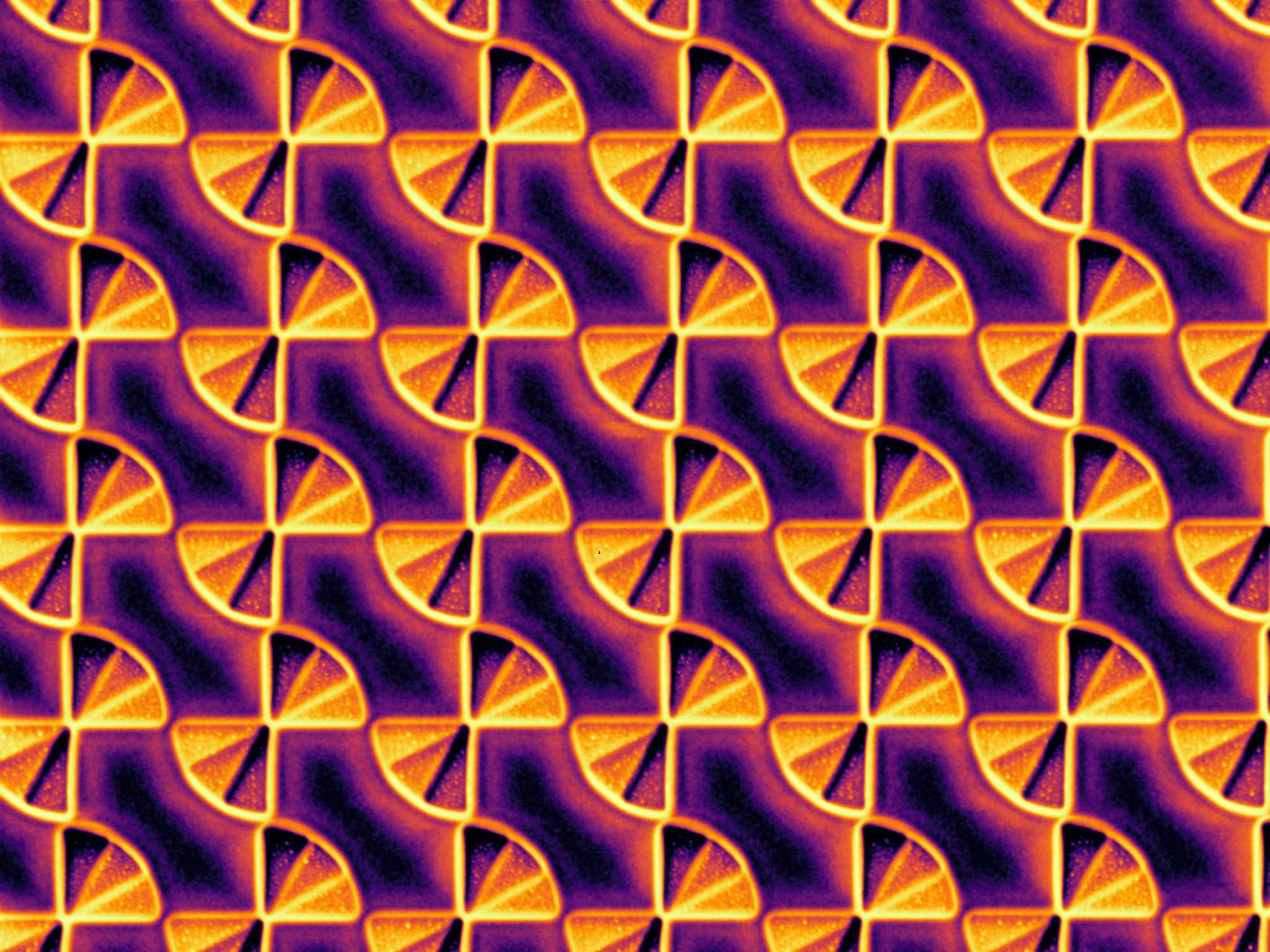
"Subwavelength Farfala" - FIB-milled infrared polarizer
This image was taken on a Zeiss NVision 40 FIB. This 2.5-D metamaterial structure would be difficult to fabricate using conventional techniques such as EBL. The stepped structure in a single-crystal hBN substrate imparts three different phase offsets to incident IR light, functionally transforming the polarization from random to left or right handed circular in a deeply subwavelength space. The whole array is 75 microns square, and the bowtie diameter is 1 micrometer and pitch is 1.2 micrometers.
Department of Chemistry & Chemical Biology, McMaster University
Supervisor: Dr. Jose Moran-Mirabal
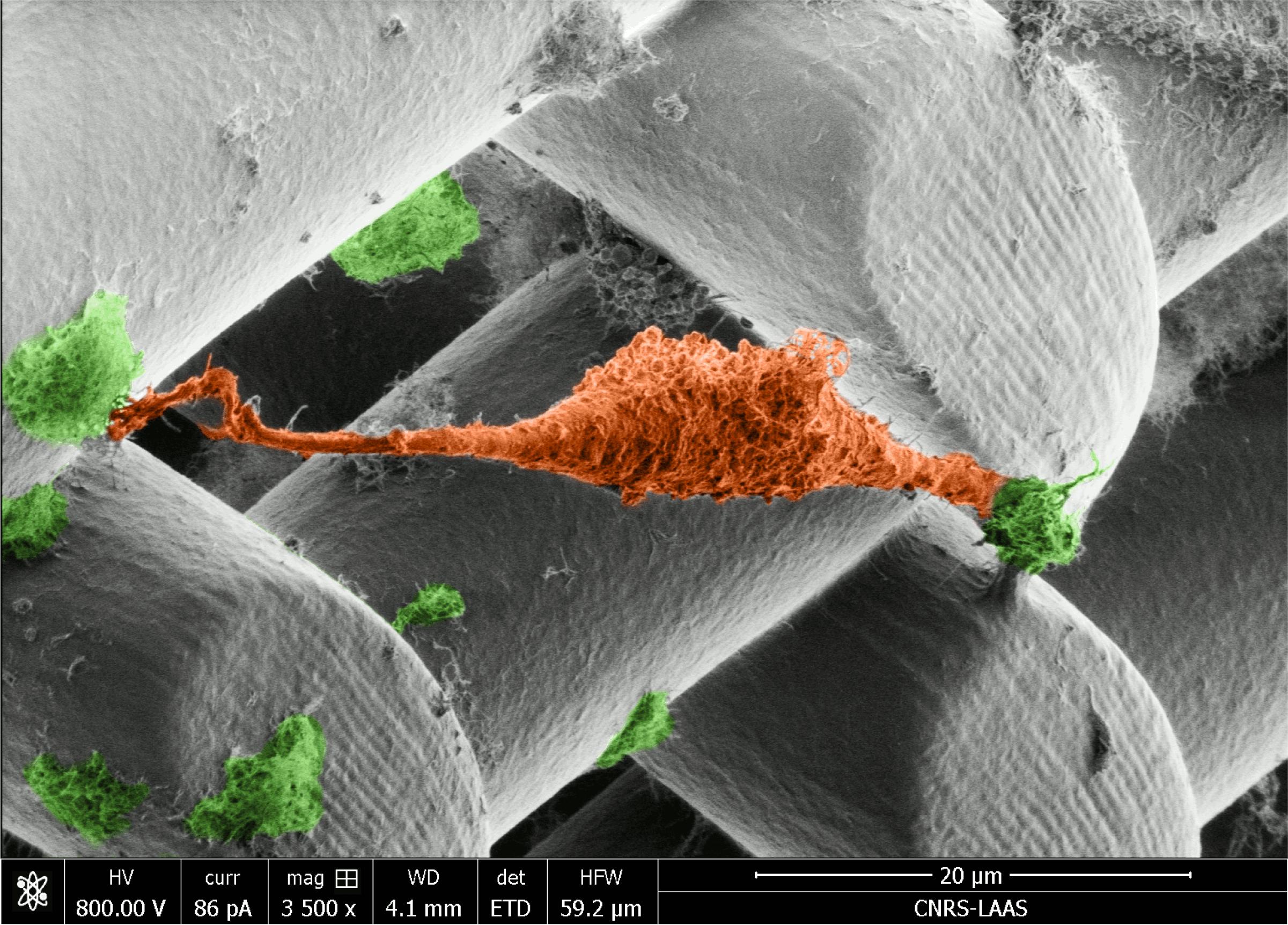
"Image acquired using EM-Helios Nanolab 600i SEM"
This micrograph shows a prostate cancer cell (orange) suspended between cellulose nanocrystal clusters (green) doped into Poly(ethylene glycol)-diacrylate (PEGDA) hydrogel scaffolds. Our work aims to develop new 3D printing inks that incorporate renewable nanomaterials. The scaffold was printed using 2-photon DLW (Nanoscribe). The Beams are 20 µm in diameter with 30 µm spacings.
Institute of Biomaterials and Biomedical Engineering (IBBME), University in Toronto
Supervisor: Dr. Warren Chan
"Bubble-flattened cells within an endothelium"
Human Umbilical Vein Endothelial Cells were seeded and grown within a microfluidic chip until they were confluent. Using fluid flow, they were subjected to shear stress, and responded by aligning their cytoskeleton fibres in the direction of flow, and were able to withstand fluid flow by expressing VE-Cadherin tight-junction proteins. During this process, however, a bubble was trapped in the chip, causing cells to flatten in a peculiar formation, as shown here. Cells were fixed and stained to identify their nuclei (blue, DAPI), actin (green, phalloidin-AF488), and VE-Cadherin proteins (red, antibody-AF555), and imaged with standard laser-scanning confocal microscopy, Plan apo 10x (Na 0.45, WD 4.0 mm 10X objective lens, Nikon A1).
Integrated Program in Neuroscience, Montreal Neurological Institute, McGill University
Supervisor: Dr. Timothy E. Kennedy
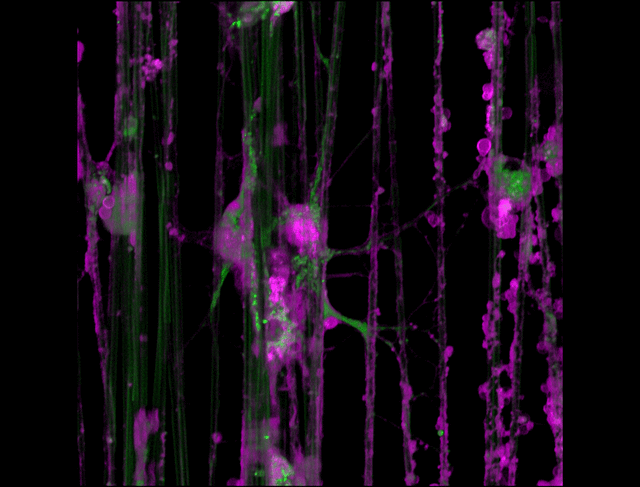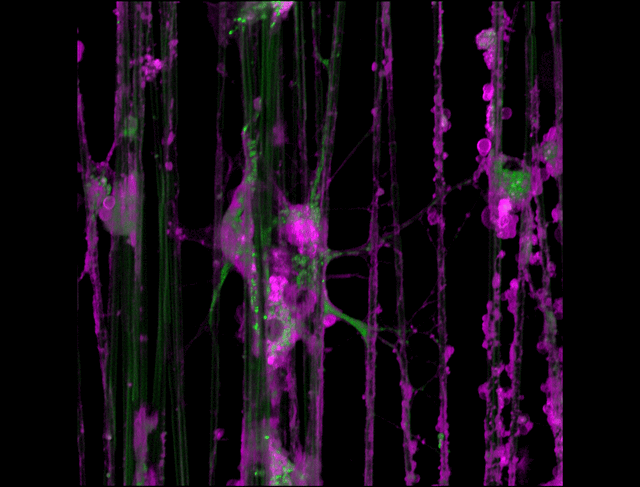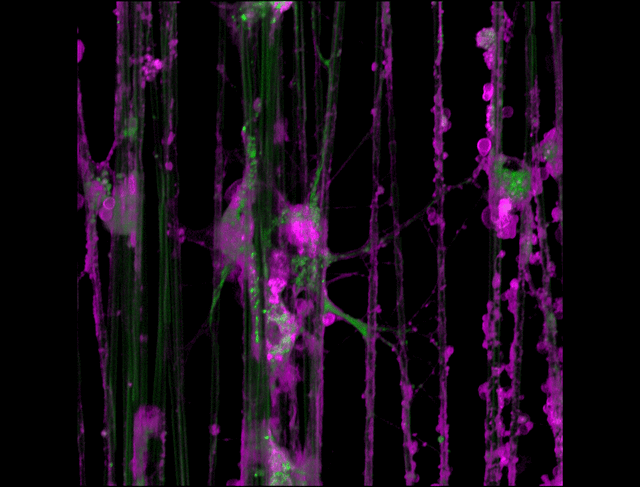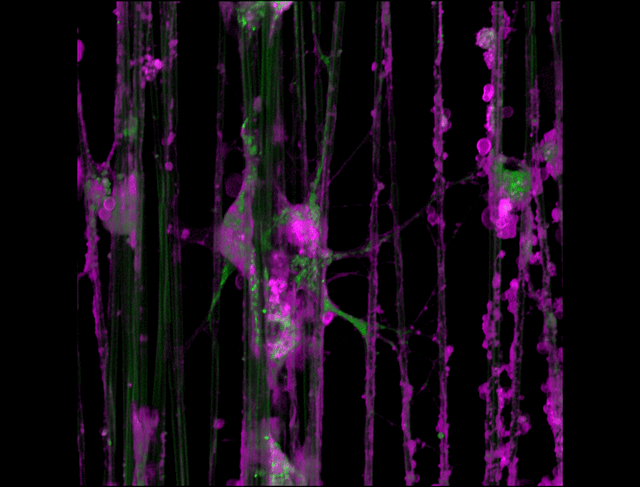
"3D timelapse of myelinating glia with different dyes"
Oligodendrocytes (OLs) are a form of glia in the central nervous system which are primarily responsible for ensheathing axons with many layers of membrane known as myelin. These myelin sheaths dramatically improve the efficiency axonal transmission by providing electrical insulation, shuttle energy sources to the axon such as lactate, and even help mediate certain forms of plasticity during learning. However, in part due to the difficulty of isolating and studying OLs, much is unknown about the cell and molecular mechanisms underlying myelination. Here, we have isolated postnatal rat OLs and cultured them for 2 weeks on axon-sized artificial fibers. As OLs differentiate, they wrap their processes around these fibers and start forming myelin sheaths as if they were axons. We used a combination of 2 fluorescent dyes – one which labels intracellular organelles (green), another which selectively binds the plasma membrane (purple). Using Airyscan Fast Acquisition Mode, a relatively new microscopy technique which allows significantly faster and higher resolution imaging than standard confocal imaging, we were able to visualize the dynamics of individual organelles, all in 3-dimensions. Imaging was performed using a Zeiss LSM880 confocal microscope with a 20X NA 0.8 objective, over 15 minutes. The incredible spatial and temporal resolution provided by this imaging technique provides an unprecedentedly clear view of how OLs produce myelin.